What is antimicrobial resistance and how does it occur?
‘Antimicrobial resistance (AMR) is resistance of a microorganism to an antimicrobial medicine to which it was previously sensitive.’ (WHO Europe)46
Some resistance may initially occur through a genetic mutation of chromosomal DNA, for example a mutation in Penicillin Binding Protein may confer resistance to beta-lactam agents.
Much of the resistance to antimicrobials, however, occurs through the presence (or acquisition) of resistance genes on mobile genetic elements (such as plasmids), the origin of which is uncertain.
Once a resistance mechanism exists in a population, it may be transferred between bacteria by both vertical and horizontal transmission.
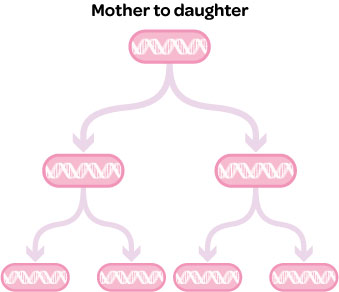 Vertical transmission
Vertical transmission
During the process of cell division, genes conferring resistance may be replicated and passed on to daughter cells.
Horizontal transmission4
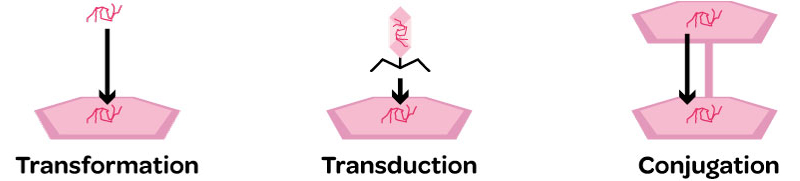
There are three methods that bacteria in a population may use to share genetic information:
1. Transformation – uptake of free DNA into the cell. Only a few species of bacteria are able to perform transduction and the DNA must be from the same or a closely related strain as the host bacterium. This is probably not important in antimicrobial resistance.
2. Transduction – delivery of a plasmid by a bacteriophage (a virus that infects bacteria). This may be used by a few species to transfer resistance genes.
3. Conjugation – transfer of plasmids between cells. This occurs during cell-to-cell contact when the host bacterium produces surface protein tubules (pili) to connect the two cells. The genes providing this ability are encoded on the conjugative plasmid, which then crosses between the two cells. Other non-conjugative plasmids can cross with a conjugative plasmid. Many Gram-negative and some Gram-positive bacteria can conjugate. This feature is often seen in dense populations of bacteria such as those in the GI tract. It usually occurs between bacteria of the same species, but some plasmids can transfer to other species by this method.
Once a resistance mechanism exists, selective pressure, for example through antimicrobial use, will cause an increase in the prevalence of this resistance in a population.
In 1944, before widespread use of penicillins, 95% of Staphylococcus aureus isolates were susceptible to penicillin; however, by 1998 this figure was 10%.47 Analysis from a study in Belfast showed that hospital incidence of extended-spectrum-beta-lactamase producing bacteria (ESBLs) had a positive relationship with use of amoxicillin-clavulanic acid in the community.48
Resistant strains may spread between individuals in both hospitals and the community.
In the UK the CTX-M family of ESBLs were first reported in 2003, and by July 2004 there were almost 500 isolates reported from more than 70 centres across England.49 The spread of resistant strains is often facilitated by poor hygiene or overcrowding: a study of children under five in a hospital in Guinea-Bissau showed that bed sharing with another child under five was a risk factor for carriage of ESBL in faecal isolates.50 Many resistant strains have now spread globally, including those with ESBLs. For example, the CTX-M-15 strain was first reported in India, and has since been found in many other countries including Canada, France and Russia.49
Disclaimer: Indications and doses may vary between products. The antimicrobials listed may constitute an off licence use of the product and as such should only be used according to the ‘Cascade’, further details of which are available on the RCVS, VMD and NOAH websites. Veterinary surgeons are advised to carefully check the Summary of Product Characteristics (SPC) before prescribing a product and obtain informed owner consent where required.

